Innovations & Thérapeutiques en Oncologie
MENUThe application of spatial transcriptomics to histological sections: a cutting-edge technique for medical diagnosis Volume 10, issue 2, March-April 2024
- Key words: spatial transcriptomics, tissues, cells, in situ hybridization, sequencing
- DOI : 10.1684/ito.2024.414
- Page(s) : 93-103
- Published in: 2024
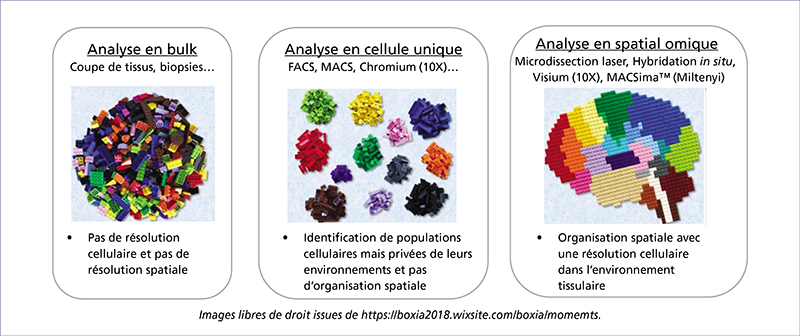
Spatial omics have been evolving rapidly in recent years, and due to the richness of the data they provide, they appear to offer opportunities for understanding biological phenomena and improving healthcare. Spatial omics enable the analysis of genomic, transcriptional, proteomic, and metabolic content at very high resolution. Beyond identifying molecular cell subpopulations, these approaches also provide information about the spatial location of the identified subpopulations within tissue, their proximity to each other, and their relationship with the extracellular matrix, blood vessels, and other tissue components.
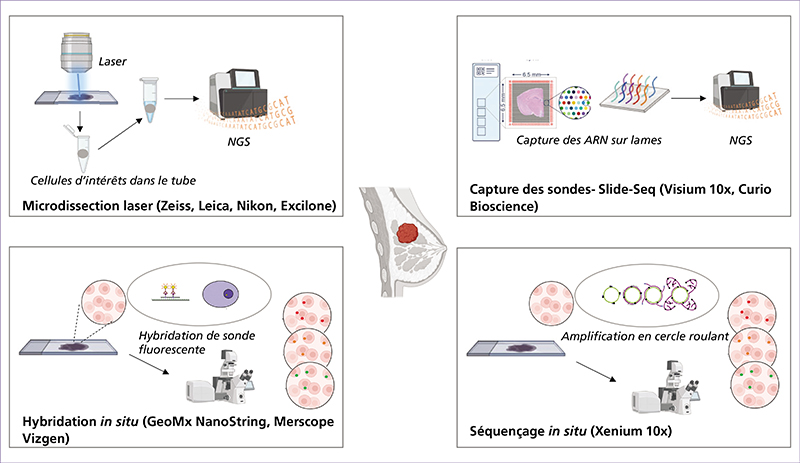
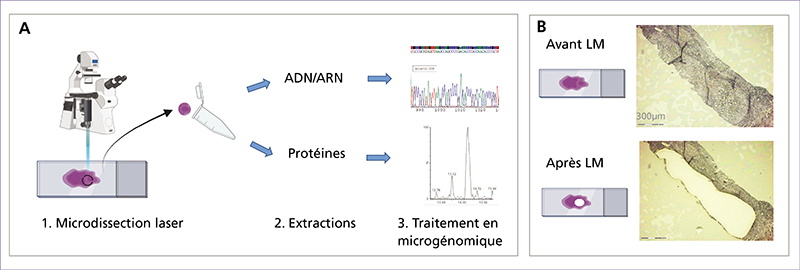
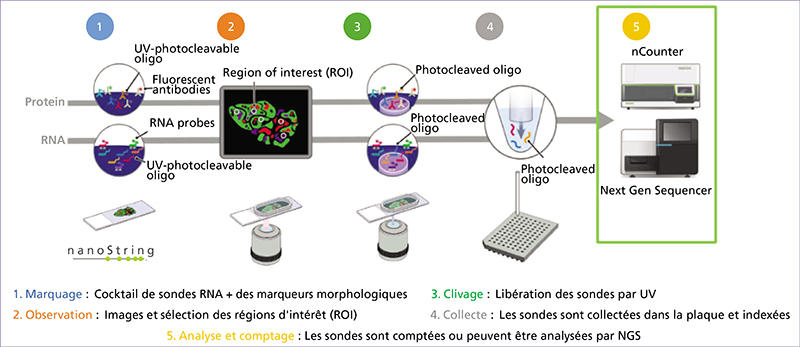
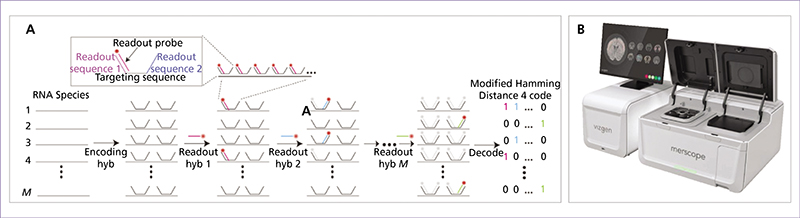
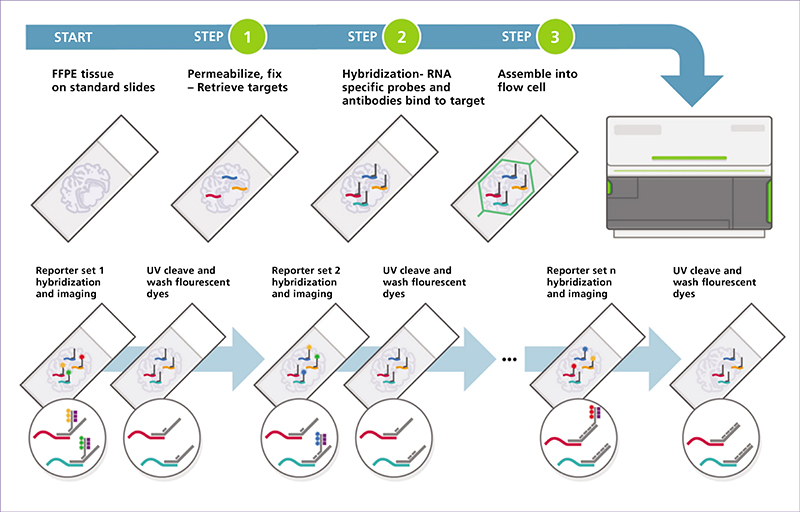
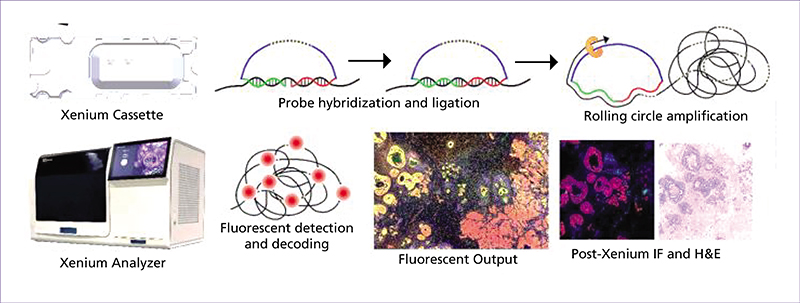
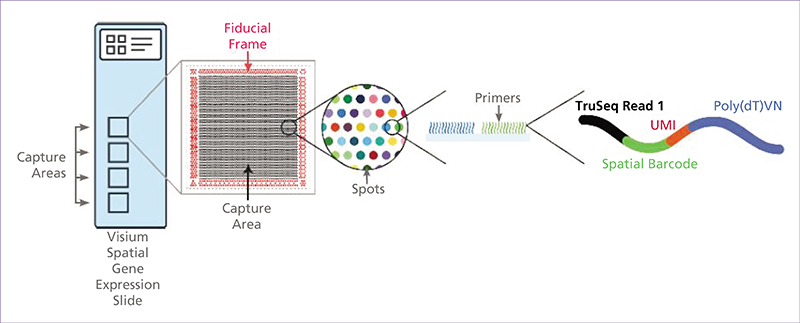
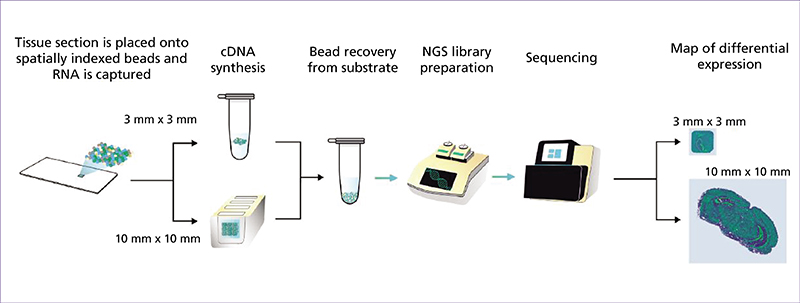
Spatial transcriptomics (ST) is an emerging research field that combines high-throughput genetic analysis with the spatial localisation of cells within tissues. Unlike traditional sequencing methods, ST offers a more comprehensive understanding of the molecular and cellular organisation of tissues. Our review explores various ST profiling strategies, including laser microdissection, which isolates specific cells from tissue for in-depth omics analyses. We then present various fluorescence in situ hybridization (FISH) techniques used in ST. Some state-of-the-art techniques allow simultaneous visualisation of numerous targeted transcripts and proteins with 3D subcellular resolution. ST techniques based on higher-resolution sequencing are also described in the final section.
Future challenges for ST include improving the spatial resolution of NGS-based methods and gaining an in-depth understanding of cellular networks and tissue interactions. The rapid evolution of these technologies offers exciting opportunities to better understand complex biological mechanisms in the spatial context of tissues.