Epileptic Disorders
MENUA web-based diagnostic reference centre for the European Reference Network “EpiCare”: recommendations of the eNeuropathology working group Volume 20, issue 5, October 2018
Global digitization continues to change medical research and practice across all specialties. Currently, pathologists in general face substantial challenges to adopt novel microscopic methods, to securitize large image repositories and to standardize analytical pipelines. Neuropathologists, in particular, are required to integrate various pre- and postoperative imaging modalities into their macroscopic, histological, and molecular reports to provide optimal guidance for antiepileptic therapies (Blümcke et al., 2017). Ideally, all these tasks, like most critical digitization efforts, should be tackled in an internationally concerted action.
Reviewing the current state of digital technology with respect to neuropathology is a necessary step towards developing guidelines within the emerging European Reference Network's epilepsy care program (ERN EpiCare, http://epi-care.eu) and beyond. Digital technology is well established across all medical disciplines and has become essential to daily routines and specialist activities alike. Whole slide imaging (WSI), affordable storage solutions, and burgeoning computation power are driving image analysis routines into the methodologic core of pathology. Undoubtedly, progress in automation and algorhythmization will bring about considerable benefits as well as novel threats to the welfare of patients and practitioners. The torrents of data created by high-throughput processes can easily overwhelm even the most conscientious diagnosticians, if sufficiently detailed methodological knowledge is not provided in the framework of postgraduate training and continuous medical education. This calls for updated curricula, detailed guidelines, and accreditation procedures encompassing the relevant aspects of digital technology to avoid jeopardizing pathology's essential function as the foundation of medical practice (Pantanowitz et al., 2013). Neuropathologists who, by default, are constantly interacting with neurophysiologists, neuroradiologists, and neurosurgeons have developed some familiarity with the data formats current in their neighbouring disciplines. The ability to integrate findings, modalities, and procedures across different fields of expertise is essential to building a truly networked version of medicine. Neuropathology is therefore ideally positioned to play a major role in shaping the multimodal pathology of the fully digitized future.
Virtual microscopy: tiled high-resolution pictures
The currently available and increasingly affordable combinations of large sensor multimegapixel sCMOS cameras, ultra-high resolution (up to 8 K) desktop monitors, and graphics processing units supporting frame rates above 60 Hz are providing detailed, jitter-free, and wide-screen visual experiences now surpassing those delivered through the eyepieces of traditional microscopes. Only long entrained habits, not picture quality, may slow the adoption of “screen scoping” in pathology diagnostics. Mere generational shift and a growing familiarity with augmented reality (AR) technology will bring about this change of pathologists’ view. Already commercially available “smart glasses” (https://tech.moverio.epson.com/en/bt-300/) can deliver live pictures of a quality sufficient for morphologic diagnosis. Just as ocular lenses, glass slides are on the retreat as the medium of long-term storage, a trend first gaining momentum in morphology-based pathology research as a growing number of automated microphotography devices sporting multiple dry and immersion objectives, motorized stages, monochrome and colour cameras and slide loaders. Virtual slides are now permeating not only pathologic diagnosis, consultation, review, publication and training, but also all morphology-based branches of the life sciences.
WSI-scanners have considerably evolved from those envisaged in the first “virtual microscopy” patents (Bacus and Bacus, 2001) and are now able to digitize standard and fluorescence microscopic sections in various formats (table 1). It currently takes under two minutes to scan conventional and special stains of a 15 × 15 mm2 section area under bright field illumination with a 20x objective; recording z-stacks and several fluorescence channels will multiply the scan times required. Individual sections are identified by their 1D- or 2D-bar coded tabs, while each partially overlapping “slide shot” is exposure-corrected and tiled by instrument-specific proprietary or open-source software in real-time or post-acquisition. Depending on tissue area, magnification, number of scanning levels, and compression formats used (non-lossy or high efficiency lossy, such as JPEG XR), the resulting image files can vary in size, reaching hundreds of GB in multi-channel fluorescence slides. Consequently, the rapid and general introduction of glass-less pathology archives even at academic centres is faced with considerable costs and the complex infrastructural demands necessary to maintain sufficiently performant retrieval and back-up logistics (Stathonikos et al., 2013).
Diverse proprietary formats vs the DICOM standard
WSI files are generally formatted in a pyramidal fashion encompassing several levels of resolution to allow continuous and rapid zooming through fields of view/regions of interests at minimized data transfer rates and RAM requirements. Unfortunately, many microscope manufacturers and biomedical software providers have developed proprietary versions of the generic pyramidal format in an attempt to entrap the user in their ecosystems (“walled gardens”) of locally installed or server-based applications. These irritant business policies in blatant disregard of the true scientific spirit have spawned a number of initiatives which develop platform-spanning alternative applications for different operating systems (Windows, Mac OS, Linux) under the GNU General Public License (GPL) model.
Exemplarily OpenSlide developed by Carnegie Mellon University School of Computer Science (http://openslide.org/) offers C libraries with Java and Python bindings to convert the WSI formats of most scanner manufacturers for viewing in standard browsers supporting HTML5 (Goode et al., 2013). Ideally, however, WSI files should comply with the “Digital Imaging and Communications in Medicine” standard, synonymous with the ISO (International Organization for Standardization) standard 12052 whose supplement 145 defines objects containing very large image data sets common to pathology (Singh et al., 2011). The creation of interoperability with the “Picture Archiving and Communications System (PACS)”, long established in most hospitals for radiology images, represents a major integrative effort which, notwithstanding the abortive attempts of a few manufacturers, has not been addressed in a coordinated fashion. Currently, most WSI are generated by brightfield instruments capable of scanning standard, special, and immunhistochemical stains as well as chromogenic in situ hybridisations (CISH); all modalities which can pose considerable challenges for downstream image analysis. More recently, the more expensive fluorescence-enabled scanners have become more popular which can provide image data for more discriminatory and reliable analyses of multi-channel immunofluorescence and fluorescence in situ hybridisations (FISH). Multispectral scanners additionally allow the unmixing of spectrally overlapping chromophores and thereby analyses of an even greater number of fluorescent markers or probes in the same section (http://www.perkinelmer.com/product/vectra-3-0-200-slide-cls142338).
Slide archiving beyond petabytes
As X-ray archiving has moved from film to PACS, pathology institutes are now en route to divest their slide cabinets for data troves of truly gigantic capacities. The resultant demands on storage media, networking infrastructure, and data security are immense. In view of the growing importance of global access to medical image for reproducible subtyping of large collectives, reference pathology of rare entities, training medical professionals, and international research communication in general investing in adequate hardware and resilient infrastructure for medical purposes should have considerable priority in global policy decisions. These issues have impact also for eNeuropathology with regard to training and agreement studies for rare epilepsy-associated brain tumours or cortical dysplasias. If medical data can be entrusted in the long term to commercial cloud services or only to institutional agencies remains to be seen given the ever-increasing exfiltration risks and criminal threats to large data storage installations of all kind. Fortunately, browser-based “open source” solutions like OMERO (http://www.openmicroscopy.org/site/products/omero) have sufficiently matured, now offering all pathology-relevant functions such as granular access rights management, platform-independent WSI-rendering, slide mark-up, and annotation on a par with their commercial alternatives (Allan et al., 2012).
Image analytical pipelines
Quite early on, WSI has been recognized as the critical enabling technology for the effective application of image analytical routines in diagnostic pathology (Fine et al., 2008). Digitized high-resolution images of large tissue areas can effect significantly increased statistical power by analysing pathological changes in their entirety or on the basis of randomly selected regions of interest (subsampling). Several commercial (www.definiens.com/solutions/diagnostics, www.visiopharm.com/solutions) and open-source (http://cellprofiler.org/; Kamentsky et al., 2011) programs for the automatic quantification of absolute or relative cell numbers, cell densities, tissue areas, and staining intensities, based on conventional stains, immunological labelling or hybridizations, are available. They provide modules for a wide range of image analytical routines, such as noise reduction and object identification which must be combined, adapted and iteratively optimized for each specific task (Valous et al., 2013); the latter procedure regularly proving the most demanding and time-consuming part of the whole process. Just as current neuropharmacological research into the efficacy of targeted immunotherapy routinely employs image analysis for quantitative histology (Sevigny et al., 2016), eventually neuropathological diagnostic practice will profitably adopt substantial parts of the toolkit already established in computational neuroscience. Naturally high-throughput metricization of even easily recognizable and well-defined morphological features, such as nuclear diameter, circumference, roundness or excentricity used for neuroglial differentiation, will exceed the patience and focus of even the most experienced pathologist (Kong et al., 2013). When even more complex algorhythms are used to derive prognostic information from conventionally or immunohistologically stained sections, the cognitive and integrative capacity of human diagnosticians is no match for image analytical pipelines (Nawaz et al., 2015; Heindl et al., 2016). Similarly, the current predominance of brightfield microscopy in routine diagnostics will have to cede ground to multichannel fluorescence which allows for more exact segmentation of cell populations and more reliable quantification of staining intensities in digitized formats.
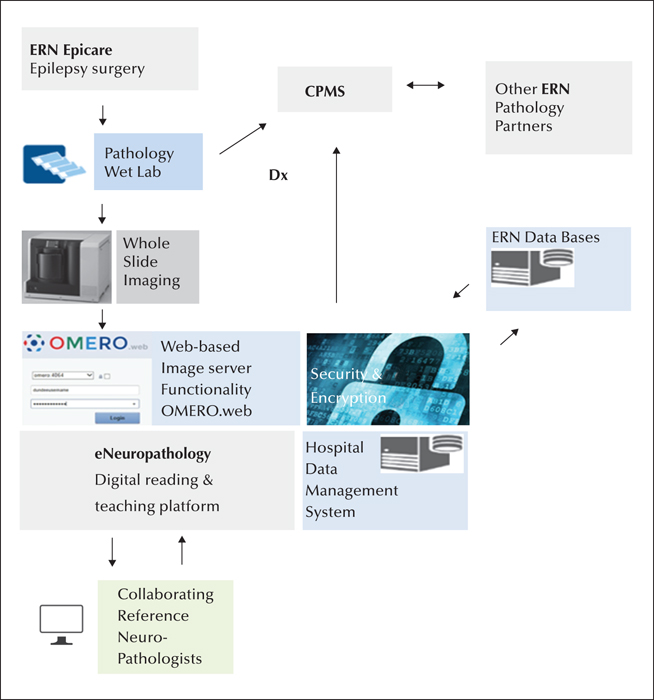
A roadmap for a diagnostic reference centre network for ERN “EpiCare”
Future navigation of the digital route towards web-based slide review, clinical audit, international agreement studies, and pathology teaching platforms will face a number of technological challenges for which technical and financial support will have to be obtained from the EU. The roadmap towards an internationally integrated European e-Neuropathology reference centre will encompass these features.
Freedom of data and software
Without doubt, every procedure in medicine should allow for its stringent and effective control. In the context of computerized applications and algorhythmic diagnostics, this requires complete transparency regarding source codes, data formats, processes, and results. Existing initiatives advocating the global availability of the results of scientific research and the preferential use of open-source over proprietary software (https://stallman.org) will eventually be difficult to oppose at least in the medical field.
Of foremost importance is the effective implementation of the DICOM Standard which has been developed with an emphasis on diagnostic medical imaging including pathology. It is already actively endorsed by the EU (https://ec.europa.eu/eip/ageing/standards/ict-and-communication/other-ict/dicom_en), however, the urgent need for its adoption needs to be impressed more forcefully on manufacturer of WSI equipment worldwide.
Integration of multiple imaging modalities
The portability and convertibility of digital data formats will enable pathologists to fuse 2D- and 3D-renditions of diagnostic samples visualized by different imaging technologies (e.g. fluorescence, CT, MRI) into multimodal composites (Magee et al., 2015).
Rapid 3D-scanning technology can provide volumetric data sets of diagnostic specimens at various stages of processing which are prerequisite to fusion images combining macro- and microscopic details. Collectively, multimodal imaging will improve morphological correlations and the interdisciplinary communication of radiological, electrophysiological, and pathological findings.
Securitization of data and networks
Local and global exchange of medical data is critically dependent on a performant network and storage infrastructure, which must be provided by the ERN hosting organization of the EU. The European Parliament has adopted the General Data Protection Regulation (GDPR EU 2016/679), that will be directly applicable in all Member States of the EU and effective from May 25th, 2018 replacing Directive 95/46/EC. Equally relevant in this context is the EU's Medical Device Regulation (EU) 2017/745 (MDR) which will be in effect on 26 May 2020. To meet the current and future e-Health requirements of European cybersecurity frameworks (http://www.consilium.europa.eu/media/31666/st14435en17.pdf), all pathology-related metadata and image files will have to be end-to-end encrypted using current state-of-the-art cryptography. Postgraduate medical education and specialist training require only network infrastructure of sufficient bandwidth, whereas internet-based image data exchange for diagnostic or review purposes needs sufficiently securitized end-to-end encryption protocols. Currently, protection from unauthorized interception can only be assured by the use of auditing the communication protocols’ source code and full disclosure of the type of cryptography used. A heightened awareness for the importance of strong encryption and its reliable implementation in all medical communications will be essential for the productive use of all types of data formats in digital pathology.
Increased performance analytics
Developments in the microscopic visualization of molecular details will reveal additional layers of complexity directly impacting on the practice of personalized medicine and theranostic capabilities of every pathologist. The recognition of morphological and mutational diversity will require a dramatic increase in diagnostic power which will only be attained in a concerted European effort to fully incorporate computerized image analysis, machine learning, and diagnostic algorhythms into practice. The need to derive prognostic information from conventionally or immunohistologically stained sections will necessitate the use of artificial intelligence (AI) techniques, such as deep neural networks (DNNs) generating complex multi-parametric decision algorithms which can match and even outperform expert-level human diagnosticians (Bhargava and Madabhushi, 2016; Djuric et al., 2017). AI-assisted pathology will only be successful, however, if accompanied by intensified efforts to standardize immunohistological staining protocols between laboratories (Lin and Chen, 2014). Several quality assurance programs in eyepiece-based diagnostic immunohistochemistry, such as NEQAS (https://ukneqas.org.uk/programmes/result/?programme=neuropathology), NordiQC (http://www.nordiqc.org/about.php) and the Canadian Immunohistochemistry Quality Control (cIQc; http://cpqa.ca/main/) are already well established.
Only immunofluorescence provides superior segmentability, signal quantitation, spatial resolution, and colocalization information to ensure the image quality mandatory for meaningful automated analyses. Therefore, future standardization efforts will have to incorporate multi-channel immunofluorescence imaging. Input into DNN will also require rich metadata encompassing methodological details on tissue fixation, processing, and staining conditions embedded in each digitized image format.
Establishing a European diagnostic reference network, i.e. eNeuropathology, will undoubtedly help in the promotion and validation of these emerging technologies across ERN neuropathology groups and accelerate their adoption in the pathology community at large (figure 1).
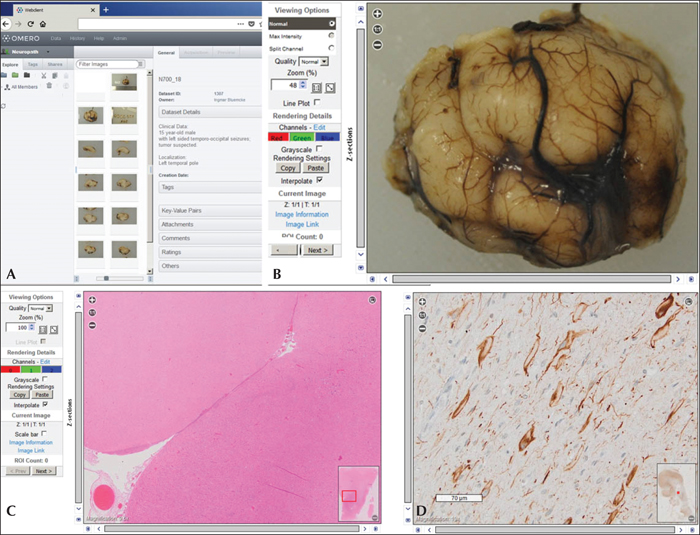
Case report (figure 2)
The open-microscopy-environment software OMERO as a platform for secure web-based neuropathological review and consultation: (A) Authenticated user's view of the OMERO landing page which lists the relevant clinical information as well as the macro- and microscopic images available for a 15-year-old male patient with left-sided focal seizures originating from the temporo-occipital lobe. (B) Macroscopic image of the surgical specimen submitted after formalin fixation showing the resected left temporal pole. The OMERO viewer allows zoomified magnification via key stroke or mouse wheel. (C) Magnification of an H&E stain demonstrating an anatomically and surgically well preserved human neocortex with inset on lower right providing whole-slide overview for orientation. (D) Immunohistochemistry using antibodies (clone SMI32) specific to the non-phosphorylated neurofilament protein, confirming the presence of enlarged dysmorphic neurons; vimentin-positive balloon cells were also identified (not shown here). The histopathological diagnosis transmitted was focal cortical dysplasia (FCD) IIb (ILAE classification 2011).
Acknowledgements and disclosures
This project has received funding from the European Union's HP-ERN-2016 European Reference Networks / Framework Partnership Agreement Grant under the Grant Agreement No 769051.
None of the authors have any conflict of interest to declare.