Innovations & Thérapeutiques en Oncologie
MENUClinical applications of next-generation sequencing in myeloid malignancies Volume 3, issue 5-6, September-December 2017
- Key words: next-generation sequencing, gene mutations, myeloid malignancies
- DOI : 10.1684/ito.2017.0099
- Page(s) : 263-70
- Published in: 2017
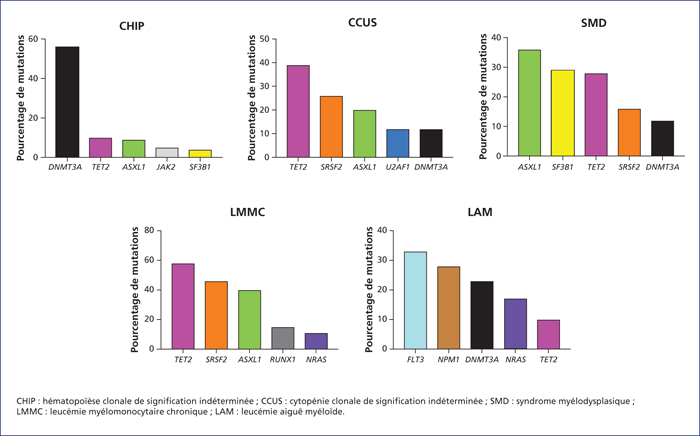
Myeloid malignancies are a heterogeneous group of diseases with a shared molecular basis. Over the last decade, the use of high-throughput sequencing has made it possible to identify recurrent gene mutations common to myelodysplastic syndromes (MDS), myeloproliferative neoplasms (MPN), and acute myeloid leukaemia (AML). These mutations now play an important role in the diagnostic process, particularly in relation to cytopenias of undetermined significance. Mutations such as those affecting NPM1, FLT3 and CEBPA in AML, SF3B1, EZH2, RUNX1 and TP53 in MDS, and ASXL1 in AML, MDS, chronic myelomonocytic leukemia (CMML), and primitive myelofibrosis (PMF) are associated with a distinct prognosis and are now routinely tested for. Beyond the mutational status itself, the extent of the mutated allele appears to influence phenotype and prognosis. Finally, in some cases, these mutations can guide therapy (when they present as potential molecular targets) or constitute molecular markers for monitoring the disease and its evolution.