Hématologie
MENUEnhancers, spatial chromosome structuring and pathological changes: towards a better understanding of complex genome alterations Ahead of print
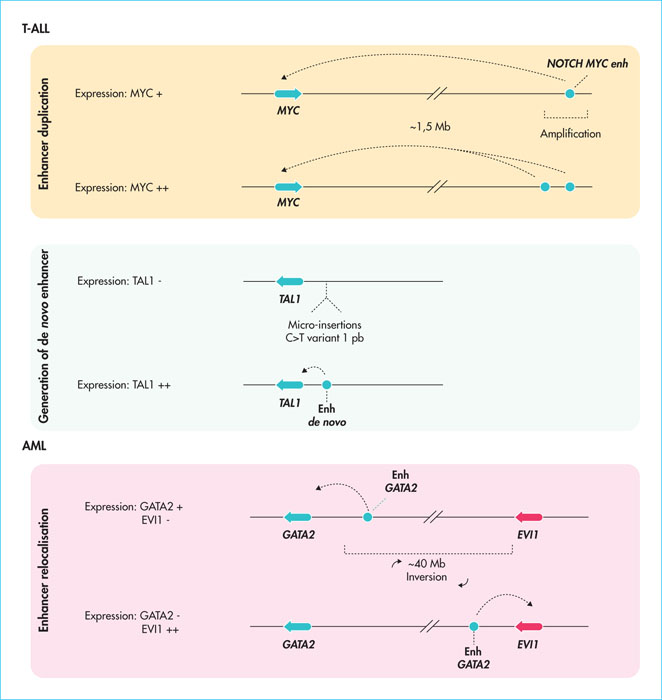
Figure 1
Examples of the deregulation of enhancers in acute leukaemia. Different cases of amplification, displacement or de novo creation of enhancers (enh) that have been implicated in the development of T-ALL or myeloid acute lymphoblastic leukaemia (AML) are represented. The result in terms of gene expression is shown for each situation.
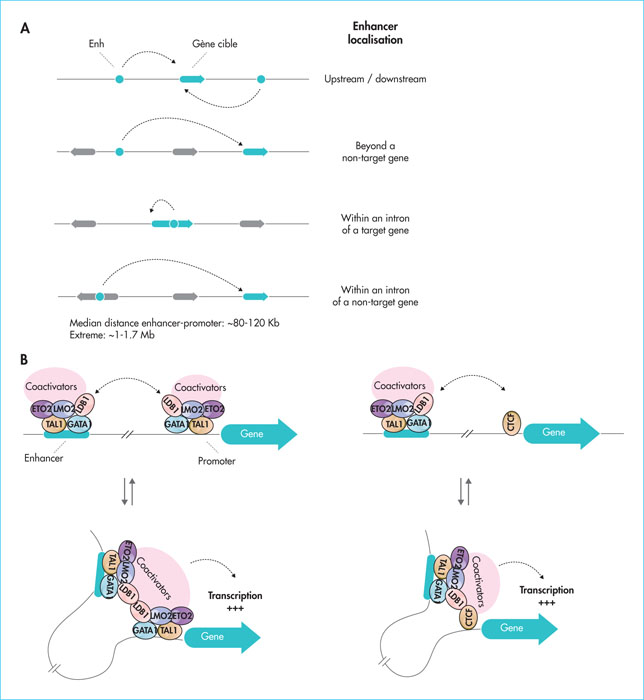
Figure 2
Localisation of enhancers in the human genome and mechanisms of control by chromatin folding. A) Enhancers can be identified upstream, downstream or within intronic sequences at distances of up to 1 Mb. B) Mechanism of control of distal enhancers by the LDB1 complex. The ability to dimerise (left side) or heterodimerise with CTCF (right side) allows LDB1 to bring enhancers close to their target genes (chromatin folding).
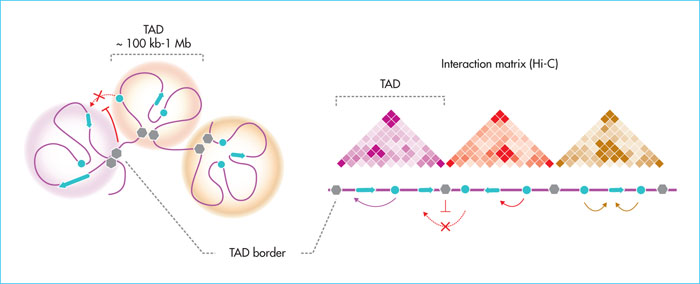
Figure 3
Topological domains (TADs) and spatial organisation of chromosomes. Theoretical schematic representation of TADs (left, spheres) containing spatially interacting regions of the genome (genes, enhancers, chromatin folding). The different TADs are separated by border regions (grey hexagons) enriched with structural factors (CTCF, cohesins) that isolate the TADs from each other. On the right, a Hi-C-type chromatin-chromatin interaction matrix corresponds to TADs. TADs promote intradomain gene-chancer interactions. The borders limit ectopic interactions between enhancers and genes in the surrounding domains.
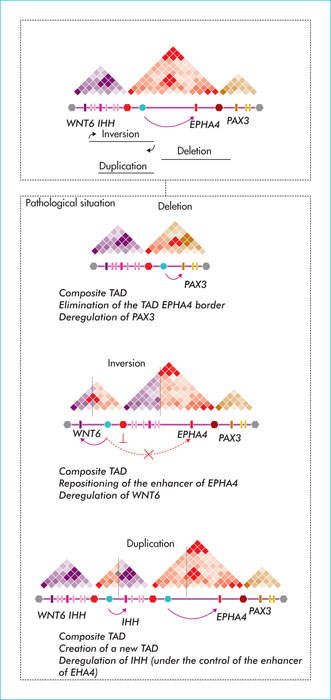
Figure 4
Pathological remodelling of TADs and spatial reorganisation of chromosomes. Chromosomal changes (inversions, duplications, deletions) involved in polydactyly identified in patients, and the spatial result at the level of the affected chromosomal regions. In each case, a border of the EPHA4 locus is moved or eliminated, causing ectopic interactions between EPHA4 and surrounding TAD genes (WNT6, PAX3).
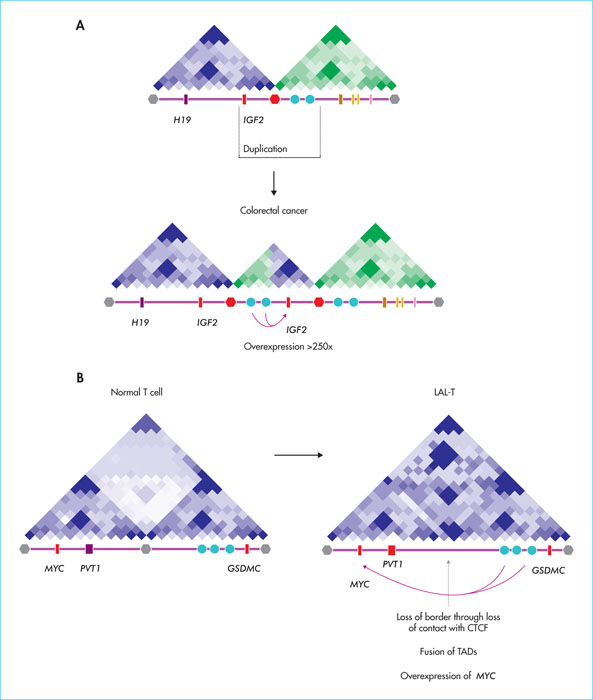
Figure 5
Remodelling of spatial organisation in cancer. A) Recurrent duplications resulting in the generation of a “neo-TAD” juxtaposing the 1GF2 gene to a super-enhancer region (blue dots), conferring overactivation of IGF2 in colorectal cancers. B) Example of TAD fusion at the MYC in T-ALL by loss of CTCF binding at the inner border, causing uncontrolled interaction of MYC with downstream enhancers. The TADs (schematic views of the Hi-C interaction matrices), the position of the genes (rectangles) as well as the borders (hexagons) are indicated.

