Epileptic Disorders
MENUThe brain as a complex network: assessment of EEG-based functional connectivity patterns in patients with childhood absence epilepsy Volume 22, issue 5, October 2020
In the past two decades, the new science of complex networks has emerged as a powerful tool to characterize the structure and function of various real-world systems with applications spanning from biological to technological, and social sciences (Barabási, 2012; Boccaletti et al., 2014; Gosak et al., 2018; Halu et al., 2019). Recent developments in the quantitative analysis of complex networks have been rapidly translated also to studies of brain network organization (Bullmore and Sporns, 2009; Park and Friston, 2013; Muldoon and Bassett, 2016; De Domenico et al., 2016). In this vein, brain areas represent nodes of a network and the edges stand either for structural connections or for functional associations. Structural brain networks usually describe anatomical parcellation of the brain and links signify physical connections between different areas that are acquired from MRI or histological data (Gong et al., 2009). An even more popular concept is the functional brain network, in which the signals from different regions or activity of sites are pairwise compared and connected on the basis of some similarity measures, such as cross-correlation (Reijneveld et al., 2007). Two nodes of the brain network are then connected if their degree of synchronization is statistically significant. Studies differ in the type of measured signal (EEG, MEG, DTI, fMRI) and protocol to build up functional connectivity maps, but they all share the same idea that the extracted brain networks are subsequently analysed with tools from the realms of the complex network theory (Rubinov and Sporns, 2010). In this manner, the brain can be reliably quantified with a small number of neurobiologically meaningful and rather easily computable measures. The idea of utilizing such advanced computational methods for assessing neurological data is nowadays established as a new discipline – network neuroscience (Bassett and Sporns, 2017; Podobnik et al., 2017; Bassett et al., 2018). These methods have demonstrated that human brain networks display properties such as a small-world and scale-free character, hierarchical modularity, and the presence of hubs, which may directly facilitate cognitive processes (Bassett and Bullmore, 2016). Most importantly, an increasing amount of evidence suggests that these characteristics are altered in disease states, thereby potentially providing important new biomarkers for neurological and psychiatric disorders (Stam, 2014; Braun et al., 2015; Navas et al., 2015; Berolt, 2019).
Construction and analysis of complex brain networks
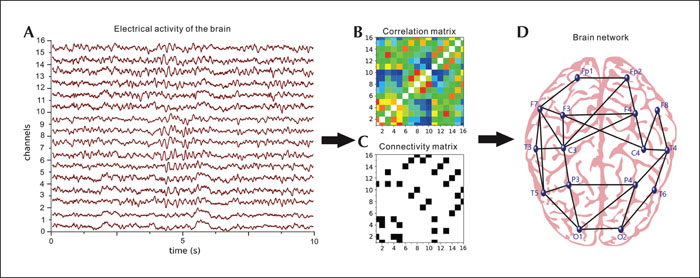
A network is a mathematical representation of a complex system, composed of two basic components — nodes (vertices) and connections, or edges, between them. Construction of a functional connectivity brain network consists of four main steps. First, one must define the network nodes, a process that depends mainly on the technique used for recording the brain activity. While in fMRI studies, parcellation into predefined anatomical regions or a single-voxel approach is typically applied, in EEG and MEG studies, the surface sensors or recording electrodes themselves can be selected as nodes (Stanley et al., 2013; Wang et al., 2010). Alternatively, EEG- or MEG-based source reconstruction techniques can be used to assess functional connectivity, however, this is computationally a very challenging task (Schoffelen and Gross, 2009; Lai et al, 2018; Anastasiadou et al, 2019). Next, irrespective of the measuring technique and the definition of nodes, a criterion for association between nodes must be established. Different analytic techniques have been developed to distinguish statistical interdependencies between two or more time series of regional activity varying from calculating the Pearson correlation coefficient to more complex methods such as quantification of synchronisation of frequencies, consistency of phase differences, and Granger causality analysis, among others (Stam and Van Straaten, 2012; Coben and Mohammad-Rezazadeh, 2015). From this, an N-by-N association matrix is generated, compiling pairwise association between all node pairs with each matrix cell representing the strength of the connection between a given pair of nodes in a graph. Generally, a threshold is applied, discarding all the links, the strength of which does not exceed a predetermined value. This produces a binary adjacency matrix, in which all the connections are considered equal (Stam, 2014). The procedure for generation of the functional brain network based on measured signals is schematically presented in figure 1.
From here, network parameters can be calculated, offering an insight into the complex neuronal architecture. The function of the brain is generally perceived as having to meet two distinct, opposing demands. Firstly, it needs to be highly segregated which enables local specialization for preforming specific tasks and secondly, it must integrate information on the global level (Reijneveld et al., 2007). Several different measures are available for quantifying various aspects of optimal brain organisation. The most important ones are described below.
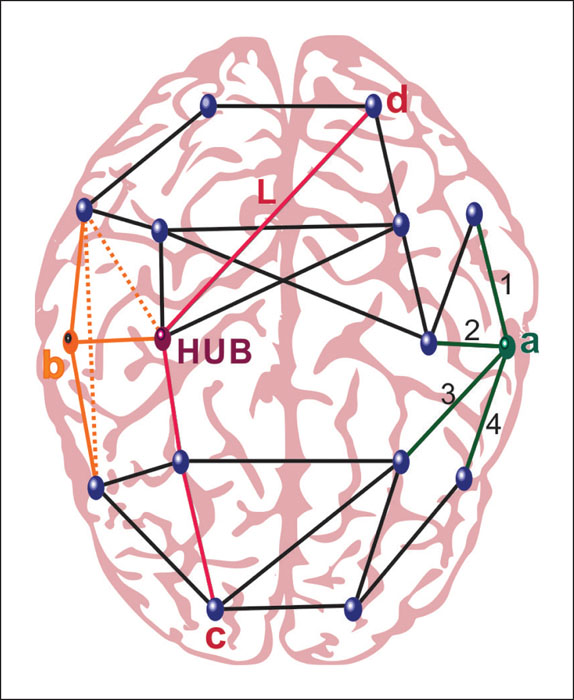
The node degree is equal to the number of connections the node forms. It is the most fundamental measure from which many others are derived upon. In terms of functional brain networks, a higher degree of a given node indicates a high level of synchronicity with many other nodes (Bullmore and Sporns, 2009). The degree distribution of a healthy brain network is heterogeneous and exhibits a small number of highly connected nodes and a high number of sparsely connected nodes. The clustering coefficient, an important parameter of local structure, is defined as a fraction of connections that are made between the neighbours of a node. In brain networks, neighbours of a node tend to be connected with each other as well, forming a cluster. In terms of function, a high level of clustering indicates high local efficiency in information transfer and resilience to random attacks and subsequent node failures (Rubinov and Sporns, 2010; Bullmore and Sporns, 2009). The shortest path length is the smallest number of edges that must be traversed to get from one node to another (Van Straaten and Stam, 2013). Brain networks have a short average path length, again supporting efficient parallel information transfer and global integration. Both of the above-mentioned attributes found in brain networks, the high level of clustering and short average path length are characteristics of the so-called “small world networks” (Watts and Strogatz, 1998; Bassett and Bullmore, 2016). As a form that is “in-between random and regular networks”, this type of organisation provides a balance between segregation and integration of information processing, and seems to be optimal for the function of many complex systems, including the brain. Furthermore, small world organization enables economic instalment of neuronal connections into the physically limited anatomical space of the skull (Bassett and Bullmore, 2016). Investigating a complex system, one is often interested in unveiling the most pivotal elements responsible for the efficient communication throughout the network. High degree nodes, which exhibit a small average distance to other nodes of the network, are considered hubs of the network and their organization is associated with inter-individual differences in cognitive performance and intelligence (Van Den Heuvel et al., 2009; Wang et al., 2010). Hubs, however, at the same time, represent weak spots since failure of a hub could have detrimental consequence on network functionality (Rubinov and Sporns, 2010). Highly connected nodes tend to be preferentially connected to other high-degree nodes, forming the so-called “rich club” found in structural as well as functional networks (Grayson et al., 2014). Brain networks are divided into modules, formed by a subset of strongly interconnected nodes with few connections to the nodes in other modules (Meunier et al., 2010). Large-scale modules, also called “communities”, in the brain network belong to the major functional system of the brain involved in specific neurological functions such as motor, somatosensory or visual areas (Stam and Van Straaten, 2012). This modular organisation is hierarchical, as smaller modules exist within larger modules (Sporns and Betzel, 2016). A schematic overview of network metrics used for the characterization of brain networks is shown in figure 2.
Brain networks in neurological diseases
Network organization in neurological disease almost always reflects a deviation from the optimal pattern, which is characterized by small-worldness, hierarchical modularity, heterogeneity, and hub nodes that are interconnected in a rich club. The extent of network changes is often correlated with the extent of the underlying structural pathology and the severity of the clinical symptoms. Already, brain networks have been studied in a plethora of altered states in neurological disorders, psychiatric states, and following injury (Stam, 2014). For example, in Alzheimer's disease, a neurodegenerative condition associated with a progressive loss of nerve cells, the functional brain networks lose their normal small-world structure, and regress towards a less efficient regular-like architecture (Supekar et al., 2008; Jalili, 2016). Moreover, previous studies indicate that hub nodes exhibit the greatest beta amyloid depositions, which leads to the conclusion that they are preferentially targeted by the disease (Dai et al., 2015). Some network analysis studies showed that network efficiency is also reduced in Parkinson's disease. In particular, Li et al. found that patients with Parkinson's disease have decreased connections in the limbic/paralimbic/subcortical module and the cognitive control/attention module (Li et al., 2017a). Moreover, in people with schizophrenia, a severe psychiatric disorder, the functional connectome displays many alterations, including reductions in putative measures of local processing, i.e. clustering coefficients, as well as increases in global integration metrics, reflected by shorter path-lengths. Consequently, schizophrenic brains have a more random-like architecture with a higher level of global integration and reduced local processing, and many of the symptoms of the disease are believed to originate from aberrantly connected networks or brain regions (Stephan et al., 2009). Other examples of studies on brain connectivity patterns include investigations of brain tumour patients, changes in brain networks in cases of autism spectrum disorders, dementia, traumatic brain injury and recovery after stroke, to name only a few (Mears and Pollard, 2016). Of course, a lot of attention has also been devoted to epileptic brain dynamics. Epilepsy is the second most common neurological disorder and in the last decade more and more studies have examined the rhythmic nature of epileptic activity from a network perspective. In the next section, we briefly summarize the main findings obtained by innovative network-based approaches in clinical epilepsy research and demonstrate a concrete example of how the EEG-derived connectivity pattern changes in cases of epileptic episodes.
EEG-based functional network analysis in epilepsy
Epilepsy is a common neurological disorder that affects approximately 1% of the world's population. It causes a hyperexcitable state of parts of the brain and is characterized by abnormal synchronized firing activity of the neurons involved in a seizure (Van Straaten and Stam, 2013). Epilepsy is increasingly recognized as a disorder of large-scale brain networks, as it is evident that otherwise healthy functional networks are recruited during epileptic activity. As seizures spread widely throughout the brain, presumably along pre-existing neural pathways, patients lose control of certain functions. These functions return when the seizure abates, implying involved brain regions are also responsible for normal brain function. What has been less clear is precisely which brain networks are involved and the extent to which functional networks are perturbed during seizures, interictal activity, and at other times (Abbott et al., 2019).
Studies suggest that epileptic brain dynamics can be described as originating from an underlying complex epileptic network that links multiple brain regions (Van Diessen et al., 2013; Van Straaten and Stam, 2013; Sargolzaei et al., 2015). Network analysis in both primary generalized and focal seizures has shown that seizures are not disorganised or chaotic events, but in fact display an organised temporal and spatial structure (Braun et al., 2015; Abbott et al., 2019). What happens with functional networks before, during and after a seizure might provide insight into the dynamic processes involved (Van Straaten and Stam, 2013). On the local level, focal epilepsy is characterized by a small brain area with abnormally increased excitability (such as the epileptogenic zone and the origin of high-frequency oscillations in temporal lobe epilepsy), increased structural connectivity (possibly the result of damage and rewiring) and one or more highly connected hubs. The local components may be responsible for the increased activity, synchronization and network regularity in the interictal state. Only if the activation exceeds a critical threshold will activity spread through general hub-like structures to the rest of the network, and this then results in a generalized seizure and a transiently hyper-regular functional network. If this process occurs repeatedly, long-distance connections and general hubs will become damaged, resulting in a loss of long-distance connectivity and, eventually, in cognitive dysfunction in patients with epilepsy (Stam, 2014).
In the following, we demonstrate how the network theory can be utilized for functional connectivity networks in six children with childhood absence epilepsy (CAE) and show how the topological features of EEG-derived networks change during epileptic episodes. The children were five to eight years of age and had normal neurological state and development. Their history was typical of brief and frequent absences without myoclonic jerks or any other type of seizure. All but one were treatment naïve at the time of EEG recording. One patient was already being treated with low-dose valproate for three days. Their EEGs showed normal background brain activity interrupted by periods of bilateral, symmetrical and synchronous discharges of 3-Hz generalised spike-and-wave (SW) discharges lasting between 4 and 18 seconds, mainly provoked by hyperventilation. Photic stimulation did not precipitate the seizures, nor subclinical discharges. Clinically, the discharges corresponded to unresponsiveness with halting of over-breathing, spontaneous eye opening and staring. There were no pronounced automatisms or myoclonia.
CAE is an idiopathic, generalized epilepsy with a typical onset between 4-10 years of age (Kessler and Mcginnis, 2019). While affected children were generally believed to have a normal neurological and cognitive development, an increased risk of attention deficit and subsequent academic difficulties has been reported (Masur et al., 2013). Clinically, frequent short-lasting absence seizures with loss of awareness and possible oral automatism in particular are observed (Matricardi et al., 2014). EEG recordings show bilaterally synchronous and symmetrical discharges of rhythmic 3-Hz spike-wave complexes with sudden onset and termination (Matricardi et al., 2014).
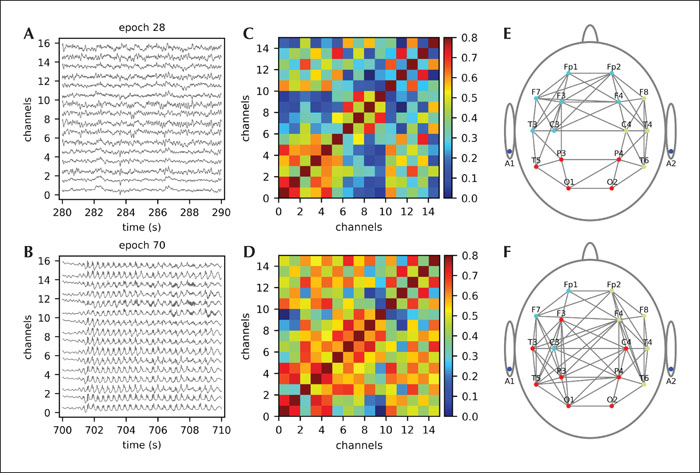
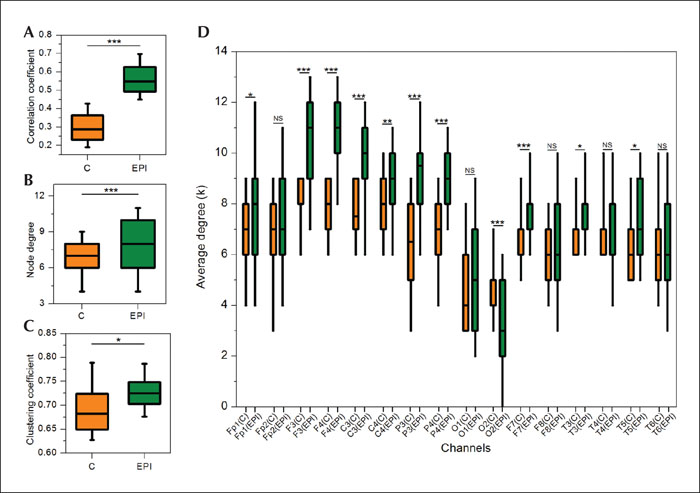
We used EEG recordings from six patients using the standard international 10-20 electrode placement. The acquisition was set to a unipolar mode, using the average of both auricular electrodes as reference sites. The patients had absence seizures during the EEG recording. The raw data was exported from the Neurofax EEG-1000 version 05-91 (Nihon Kohden, Shinjuku-ku, Japan) program used in the University Medical Centre Maribor in the ASCI format and subsequently analysed using custom-made Python scripts. The study was performed with written consent obtained from subjects’ parents/caregivers and with the approval of the Ethical committee of University Medical Centre Maribor (UKC-MB-KME-31/20). First, the EEG signals were band-pass filtered between 0.1 and 70.0 Hz. Sampling frequency was 512 Hz. The approximate 20-minute recordings were divided into 10-second epochs and altogether 60 artefact-free epochs were chosen for further analysis, 30 from normal seizure-free brain activity (control epochs; C) and 30 including generalized SW discharges (epileptic epochs; EPI), corresponding clinically to absence seizures. In figure 3, the procedure of EEG-functional network construction is presented for a typical epoch with control (figure 3A) and a typical epoch with epileptic (figure 3B) activity. To evaluate the synchronicity of the recorded signals, we calculated the pairwise cross-correlation between all traces of a recording. Correlation matrices were established for each epoch and thresholds were determined for control and epileptic epochs, respectively (figure 3C, D), based on the average value Ravg(i) and standard deviation RSD(i) of the correlation matrix for the i-th epoch. In particular, the connectivity threshold RTH(i) for the i-th epoch was then determined as RTH(i)= Ravg(i)+0.1·RSD(i). Finally, from the correlation matrices, functional connectivity networks were constructed (figure 3D, E). Comparison of both connectivity maps indicates denser networks during epileptic seizures. In order to gain more precise insight into the topological reorganization during the epileptic episodes and quantify the changes seen, some network metrics were calculated based on all 30 epochs in the given group. The box-plots in figure 4A, B, C show the average correlation coefficients, node degrees and clustering coefficients, respectively. Evidently, time series of seizure-free periods (C) are much less synchronized and their corresponding networks are less dense and also less locally clustered when compared to networks from epileptic periods (EPI). This corroborates previous studies (Braun et al., 2015; Kramer and Cash, 2012; Stam, 2014). Additionally, we evaluated the differences in node degrees of individual channels in order to gain some insight into the role of individual brain areas during epileptic activity (figure 4D). The node degree of almost all channels increased significantly during epileptic activity with the exception of channels Fp2, O1, F8, T4 and T6, where the change was insignificant, and channel O2, where the average number of connections decreased. For a more in-depth investigation of epileptic activity initiation and propagation, more advanced theoretical approaches with additional imaging techniques should be applied. However, despite the low spatial resolution of EEG recordings, this simple analysis of routinely available EEG recordings could offer crude insight into topological changes of network dynamics during epileptic activity. The ease of data acquisition and the simplicity of the analysis makes this a promising approach that could help clinicians in their daily practice.
Multilayer epileptic brain networks
Recent research suggests that the standard network approach might be an over-simplification (Boccaletti et al., 2014; Kivelä et al., 2014; Aleta and Moreno, 2019). Namely, brain network construction is liable to aggregation, averaging or disregarding a certain portion of the recorded data, mainly due to limitations in recording equipment and mathematical analytical tools available. This inevitably leads to loss of some crucial information about brain functional connectivity. An elegant solution to the problem emerged with the development of a multilayer network approach. A multilayer network can be thought of as a network of networks or a collection of inter-connected networks, i.e. each network offers a specific type of information about the brain and simultaneously acknowledges interlayer connectivity (Bassett and Sporns, 2017; De Domenico, 2017). Most often, this formalism is used to present connectivity in different frequency bands and variability of connectivity over different time scales or with respect to different task execution, and enlighten the relationship between structural and functional connections of the brain (Muldoon and Bassett, 2016; Gosak et al., 2018; Vaiana and Muldoon, 2018). While the methodology is still being developed and improved, its application to brain functional data has already uncovered several interesting features, from the emergence of new interlayer hubs and their significance in neuropsychiatric disease to the importance of network reconfigurations during the resting state on a millisecond scale, to name just a few (De Domenico et al., 2016; Kabbara et al., 2017). Noteworthy, the utilization of multilayer network formalism is recently gaining attention also in the context of epileptic brain networks (Yu et al., 2020).
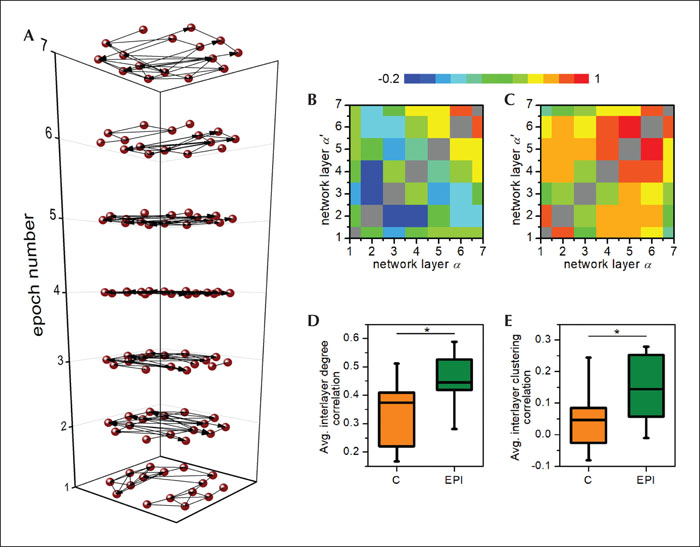
Here, we present an example of how the multilayer concepts can be used to identify different kinds of changes in the brain's collective activity during epileptic episodes. For this purpose, we took all possible subsequent two-second epochs from epileptic activity intervals from all patients, which lasted 12 seconds or more, in order to have at least six subsequent epochs for interlayer analysis. In this manner, we derived 11 series of six to eight epileptic epochs from four different patients and 11 equivalent series of artefact-free epochs of control activity from the same four patients. Then, instead of averaging the extracted correlation matrices prior to the network construction, we designed a network layer for each epoch separately, as visualized in figure 5A. This enabled us to assess the dynamic persistency of the functional connectivity patterns and to investigate how this changes during epileptic activity compared to control conditions. In particular, we calculated the Pearson correlation coefficient between the α-th and α’-th network layer (Boccaletti et al., 2014) separately for the degree and clustering interlayer correlations. To account for different average degrees, we used a variable connectivity threshold and set the average degree to five in all network layers. The matrices showing the degree of interlayer degree correlations between subsequent network layers for the normal and epileptic episodes are shown in figure 5B, C. Evidently, the correlations are higher for networks corresponding to epileptic activity. In figure 5D, E, we show the average values for interlayer degree and clustering correlations for all 11 epoch series for the normal and epileptic activity. Our results indicate that the EEG-derived brain network during the resting state is rather dynamic, whereas during the phases of absence seizures, the spatio-temporal activity and the resulting network topology are less variable. It should be noted that this is just a demonstration on how the multilayer network formalism can be used to quantify the collective activity of the brain. Further studies will be necessary to explore how the persistency of the network structure depends on the epoch lengths, frequency bands, and, of course, type of epileptic seizure. The multilayer brain network approach also offers the potential to examine the temporal evolution and dynamics of epileptic brain networks, which can lead to important implications for studying dynamic interactions during pre-ictal, ictal, and post-ictal periods (Yaffe et al., 2015), and by tracking subtle differences in network dynamics, possibly also to the detection of seizure onsets before they become evident electrographically. Moreover, the proposed formalism could provide a suitable theoretical framework to examine the evolution of brain networks over much longer periods of time, such as in EEG long-term monitoring or even to track the network reconfigurations of the same patient over years.
Discussion and conclusion
The EEG provides a fundamental tool in the primary diagnosis of epilepsy. It supports the notion of characteristic temporal events, such as interictal spikes associated with epileptic foci, and allows one to distinguish between generalized and focal neurophysiological correlates of epilepsy (Rosenow et al., 2015). However, an interpretation of EEG recordings based solely on visual inspection is very subjective and prone to human error (Kirmani, 2013). In recent years, many efforts have been devoted to the development of automated techniques (Cabrerizo et al., 2011). The success of these methods depends heavily on the number of extracted parameters, and the functional connectivity networks have proven to be a valuable repertoire that can significantly improve the precision of such algorithms (Sargolzaei et al., 2015).
Moreover, EEG-derived brain networks were not only found to be beneficial for the epilepsy diagnosis but also for its treatment. The ability to identify the seizure onset zone through the computation of network diagnostics has important clinical implications, as this could improve the localisation of brain areas that are appropriate for resection, revealing candidates for surgical treatment among patients with drug-resistant epilepsy (Braun et al., 2015). Wilke et al. studied whether critical nodes could be identified in the networks of patients undergoing epilepsy surgery (Wilke et al., 2011). Using EEG recordings, they found that a reduced number of seizures post-surgery was associated with resection of brain regions that had the highest betweenness centrality, also suggesting that critical network points are involved in either the start or spreading of seizures. In a prospective study of individuals with both brain tumours and epilepsy by van Dellen et al., the networks of individuals who became seizure-free after surgery were more integrated and showed higher centrality at follow-up than the networks of patients who were not seizure-free after surgery (Van Dellen et al., 2014).
Several of the above-mentioned aspects of network theory utilization have already been investigated in CAE – from the perspective of both structural as well as functional connectivity. This research has provided an important foundation for better understanding the initiation and spreading of absence seizures (Bear et al., 2019). The structural networks in CAE show several deviations from the optimal complex system organization, such as a decrease in small-worldness scalar on the global level as well as decreased connectivity and efficiency of specific subnetworks (Xue et al., 2014; Qiu et al., 2017). Even more interesting are the changes in functional connectivity that confirm well known facts, such as the crucial role cortico-thalamic connectivity plays in CEA (Li et al., 2017b; Jiang et al., 2019). On the other hand, this new approach provides new insight into the development of seizures in CAE. In contrast to the general perception of SW discharges occurring abruptly as bilaterally synchronous generalized events, a detailed network approach to spatial and temporal profiles of seizure development showed a low-frequency frontal cortical source preceded by an occipital source prior to the first SW discharges (Gupta et al., 2011). Critical hubs in focal cortical, subcortical and cerebellar regions during seizures were identified, likely involved in seizure generation and/or maintenance (Youssofzadeh et al., 2018). Investigations devoted explicitly to the genesis of the generalised hyper-synchronous SW discharges during seizures have revealed that the transition of SW discharge pattern from cortical local generation to generalization does not occur symmetrically, but is heterogeneous and exhibits dynamic time lags (Amor et al., 2009; Sarrigiannis et al., 2018). This might be related to our observation that the changes in time-averaged network structure occurring during seizures were not symmetric, as the node degree was increased more profoundly in the left hemisphere (see figure 4D). Furthermore, seizure termination was found to be a gradual process in which several cortical, particularly frontal, areas are involved (Jiang et al., 2019). Noteworthy, a study that focused solely on background brain activity in patients suffering from absence seizures revealed that the alpha-band functional network profiles exhibit a higher inter-module connectivity in comparison to those from healthy subjects, which implies the facilitation of emerging epileptic discharges (Chavez et al., 2010).
Despite an increasing amount of data describing changes in network parameters in patients with CEA, the link between these parameters and clinical application remains elusive. Currently, it would appear that altered functional connectivity may help in better understanding associated cognitive comorbidities (Bear et al., 2019). Functionally decreased connectivity and deactivation in default network mode were found both in conjunction with generalised SW activity and during the interictal period, and these abnormalities in the default mode network could be related to cognitive impairment during seizures (Laufs et al., 2006; Luo et al., 2011). At the same time, new network measures, such as the connection coefficient, are being developed to detect and characterize ictal states in CEA which would ultimately allow seizures to be detected automatically (Giudice et al., 2017). A potential for clinical use also became clear when different focal areas with a high degree of local connectivity identified in the narrow pre-ictal temporal window were found to be predictive of treatment responsiveness in patients with CAE, as were the characteristics of ictal networks before treatment (Tenney et al., 2018; Ossenblok et al., 2019).
It should be noted that scalp-level EEG analysis, as demonstrated in the present paper, does not allow direct interpretations in terms of underlying neuroanatomy. Accordingly, many efforts were made to reconstruct active brain sources from scalp signals. However, the signals detected by each electrode result not only from the underlying neurons, but from all active sources, superposed as a function of their distance and orientation. This makes the reconstruction a computationally very demanding problem that is additionally prone to spurious results due to volume conduction and the effects of field spread (Schoffelen and Gross, 2009; Anastasiadou et al., 2019). For these reasons, EEG-based network analyses based on the level of reconstructed sources are not that common in clinical practice. In contrast, scalp EEG brain networks are relatively straightforward to derive and are therefore the most common approach despite the limited neurobiological interpretation, including EEG brain functional connectivity networks in paediatric epilepsy (Sargolzaei et al., 2015). However, caution is required upon investigation of scalp-EEG networks, since spurious estimates of functional connectivity can occur between the channels due to the effects of volume conduction and therefore often leakage corrections are desirable (Lai et al, 2018). Moreover, the susceptibility to produce such spurious data and the interpretation of the network metrics can also depend on the choice of the recording reference and correlation metrics (Anastasiadou et al., 2019).
To conclude, the utilization of network approaches to study the collective activity of the healthy and diseased brain is a rapidly developing field and one of the hottest topics in the neuroscientific community (Bassett and Sporns, 2017; Lynn and Bassett, 2018). In the last decade, brain connectivity concepts are becoming increasingly important also in terms of clinical applications (Stam, 2014; Sargolzaei et al., 2015; Afshari and Jalili, 2017). Particularly in the field of epilepsy, EEG-based connectivity maps have gained significant prominence in the assessment of brain function with the potential to provide a decision support system for epilepsy diagnosis and seizure prediction and treatment. In the present contribution, we aimed to introduce the clinical neurologist and epileptologist to this new theoretical paradigm and demonstrate a concrete example of how networks are constructed from real clinical EEG data. We believe that such interdisciplinary endeavours have sufficiently matured to be able to start to address the many challenges of our time, not least aiding the diagnosis and treatment of disease, even though there remains significant challenges before such approaches are used in everyday practice.
Supplementary data
Summary didactic slides are available on the www.epilepticdisorders.com website.
Acknowledgements and disclosures
The authors acknowledge the financial support from the Slovenian Research Agency (Research Core Funding No. P3-0396).
None of the authors have any conflict of interest to declare.
![]() This work is licensed under a
Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License
This work is licensed under a
Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License