Epileptic Disorders
MENUEvaluation of candidate genes in a Chinese cohort of atypical Rolandic epilepsy Volume 23, issue 4, August 2021
- Key words: atypical Rolandic epilepsy, candidate genes, whole-exome sequencing, PRRT2
- DOI : 10.1684/epd.2021.1308
- Page(s) : 623-32
- Published in: 2021
Objective. We aimed to identify new candidate pathogenic genes for atypical Rolandic epilepsy.
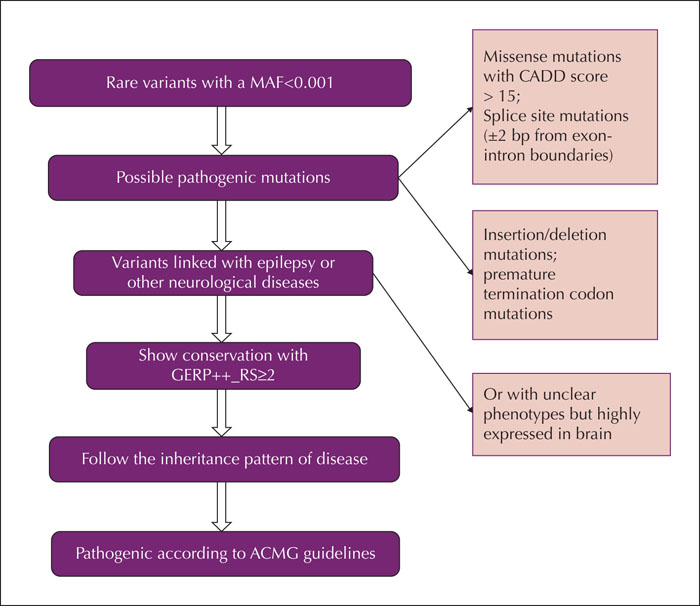
Methods. We retrospectively evaluated the data from 24 Chinese patients with atypical Rolandic epilepsy who underwent whole-exome sequencing. Data were analysed regarding the frequency of affected genes, previously reported disease-related genes, and evaluation based on Kyoto Encyclopaedia of Genes and Genomes (KEGG).
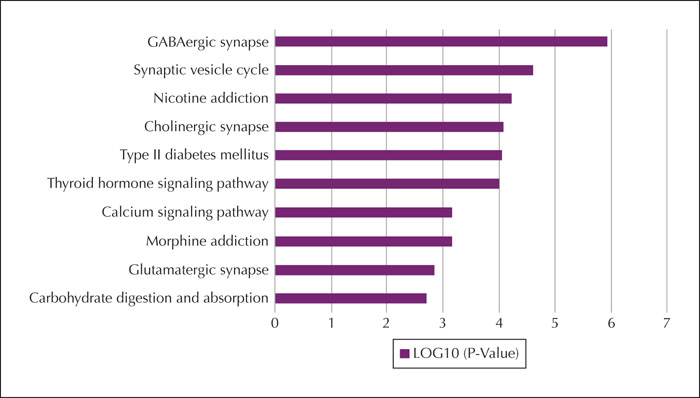
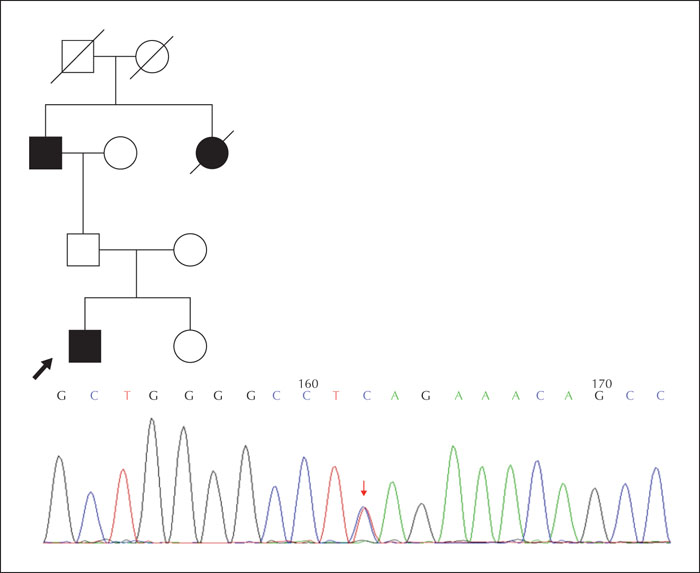
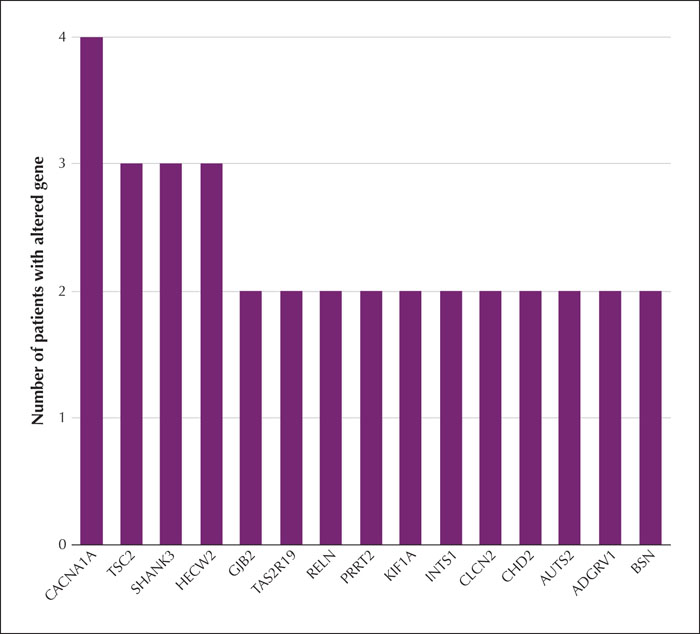
Results.We identified a frameshift mutation in the reported gene PRRT2, which is classified as pathogenic according to American College of Medical Genetics and Genomics guidelines (ACMG). We also identified a novel missense mutation in the PRRT2 gene in a family with three affected patients. Several other candidate genes were found in at least two patients, some of which were associated with other epilepsies (ADGRV1, CACNA1A, CHD2, CLCN2, HECW2, KIF1A, NPRL3, RELN and TSC2), while others were mainly associated with neuropsychiatric disease (SHANK3 and AUTS2). The KEGG analysis of 81 candidate genes associated with atypical Rolandic epilepsy identified a significant association with the GABAergic synapse. Candidate genes involved in the GABAergic synapse pathway included NSF, CACNA1A, as well as others.
Significance. Our study indicates that PRRT2 mutations may be associated with atypical Rolandic epilepsy. Moreover, we identified a number of unreported candidate genes, including ADGRV1, CHD2, CACNA1A, NSF, NPRL3, KIF1A, GJB2 and HECW2, also associated with atypical Rolandic epilepsy.